O-GlcNAc and other dynamic protein modifications
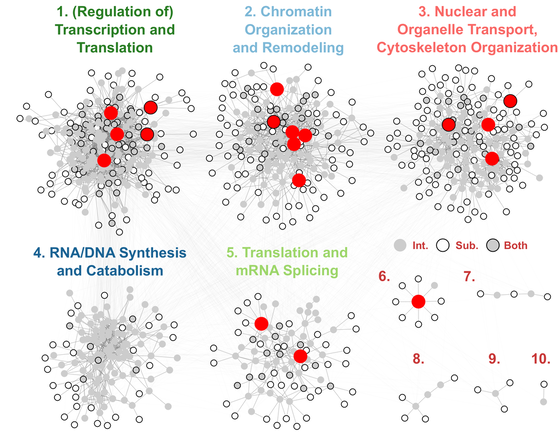
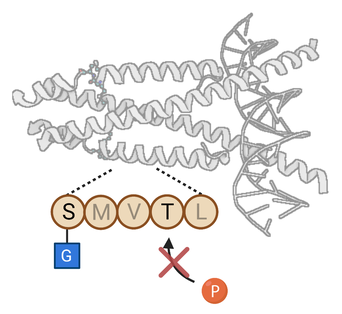
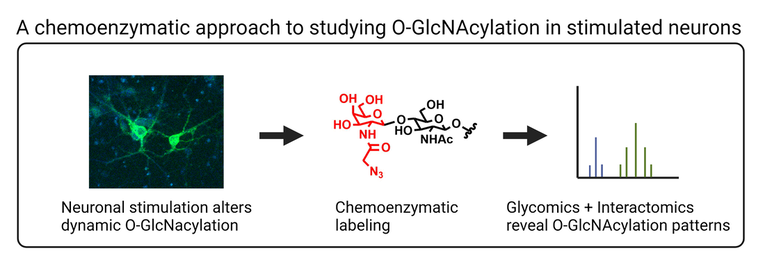
The post-translational modification of proteins is a fundamental mechanism by which information is transmitted and stored within the cell. Our lab studies O-GlcNAc glycosylation, the covalent attachment of N-acetylglucosamine to serine and threonine residues of proteins. O-GlcNAcylation is a reversible and intracellular form of glycosylation, sharing some similarities with protein phosphorylation. O-GlcNAc is essential for cell survival and plays important roles in many biological processes (e.g., transcription, translation, cell division) and human diseases (e.g., diabetes, Alzheimer’s disease, cancer).