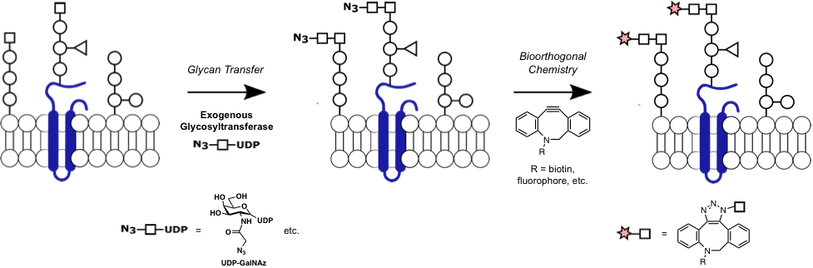
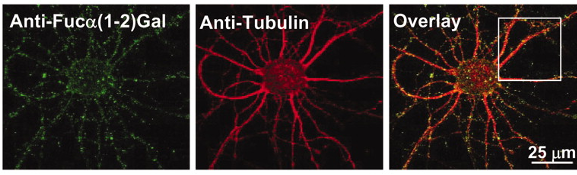
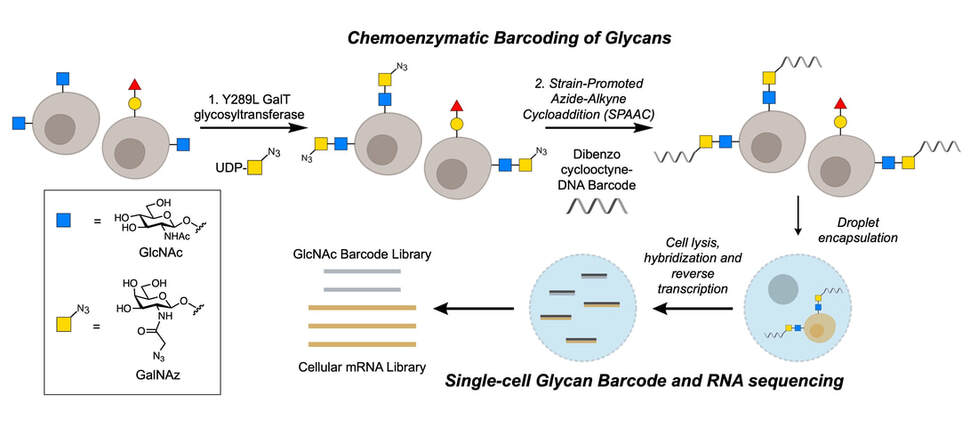
Glycan structure identification at the single-cell level
The brain is a highly complex organ, comprised of many different cell types and subclasses of cells. Our understanding of this cellular complexity has rapidly expanded with the advent of single-cell methods such as single-cell RNA sequencing (scRNA-seq). Single cell methods have been powerful in advancing our understanding of genetic molecular signatures, however, analogous methods to examine the glycan molecular signature of single cells have been lacking. Our lab has expanded our chemoenzymatic labeling strategies to specifically detect and profile the glycan signature of individual cells. In multiplexing with scRNA-seq, thus classifying cells by both gene expression and glycan expression profiles, we identify specific glycan structures, biosynthesis enzymes, glycoproteins, binding proteins, and cell types that contribute to brain processes such as learning, memory, and disease.